ModMED: Modelling Mediterrean Ecosystem Dynamics
 ModMED was an international ecological research project studying vegetation dynamics in semi-natural vegetation of the Mediterranean region. The objective was to understand the behaviour of vegetation and to be able to construct predictive models that can be used in thefuture management of burning and grazing.
ModMED was an international ecological research project studying vegetation dynamics in semi-natural vegetation of the Mediterranean region. The objective was to understand the behaviour of vegetation and to be able to construct predictive models that can be used in thefuture management of burning and grazing.
ModMED was based on a GIS-based modelling system, for handling landscape-level processes such as water movement and seed dispersal, combined with a variety of alternative vegetation models at the patch level. Simile was used for developing the vegetation models.
Recent developments in agricultural policy and land use have resulted in major changes in the ecology of Mediterranean ecosystems. Changes in grazing patterns and fire frequency and intensity have been major contributing factors.
The ModMED project is concerned with the dynamics of vegetation in response to these changes. The relevant ecological processes at the landscape, community and individual levels are being researched in Italy, Portugal and Greece and represented in a hierarchical, modular modelling environment.
The ModMED modelling environment provides a powerful tool for scientific research into the ecology and management of semi-natural vegetation, and for decision making in resource management at regional and national levels. Simile has played an important role in developing models at the community and individual levels, for use by themselves and for incorporation into Landlord, ModMED’s landscape-level modelling framework.
The following sections illustrate some of the uses of Simile within ModMED. The examples are:
- a simple model of photosynthesis;
- a model of crown development, with the crown divided into horizontal layers and radial segments;
- a detailed model of multiple-species vegetation dynamics; and
- the integration of a model of seed production and dispersal in the landscape-level modelling framework.
For more information on the role of Simile in ModMED modelling, please consult the following two documents, which were extracted from the ModMED Final Report:
- Community level models (available as pdf, 1.62 MB)
- Integration between landscape and community models (available as pdf, 1.18 MB)
Please note that many people were involved in the ModMED project. A list of the principal participants is included in both of the above documents.
More information on ModMED can be found on the ModMED web site
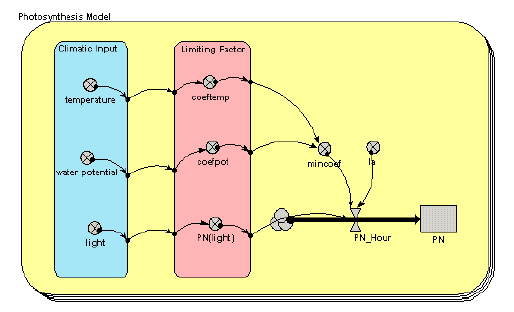
Photosynthesis model
This simple model uses three climatic inputs (temperature, water potential and light) to calculate net photosynthesis for each of several species. Note the use of a multiple-instance submodel (the round-cornered box, with multiple boundaries), with one instance for each species.
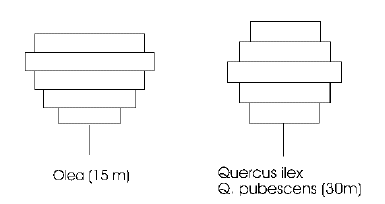
Tree crown model
This model represents the canopy of each tree as a series of layers, with each layer divided into 16 segments. The layers allow typical species-specific crown profiles to develop: the two examples shown here are for olive and oak. The segments allow the horizontal extension of the canopy to be limited when it encounters a neighbouring tree.
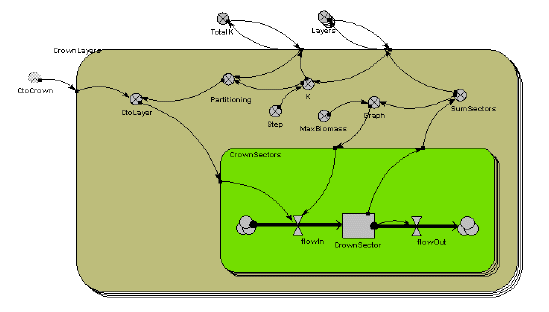
Below is the Simile model diagram for the crown of a single tree. You can see that it consists of two, nested, multiple-instance submodels. The outer submodel is for each crown layer. The inner submodel is for the 16 segments in each layer.
The crown model was in fact part of a larger, stand-level model, representing a collection of trees. In addition, the roots were also modelled, in terms of a series of root discs. In order to visualise the dynamics of the complete model, the Naples team that developed the model also developed a specialised display tool. This produces output in a 3D language called VRML, which allows the user to rotate, zoom and pan the image interactively. The following two screen shots show two views of the modelled stand of trees at some stage during the simulation, viewed from above and near ground level.

The root discs can be clearly seen in the latter view.

N-species vegetation dynamics
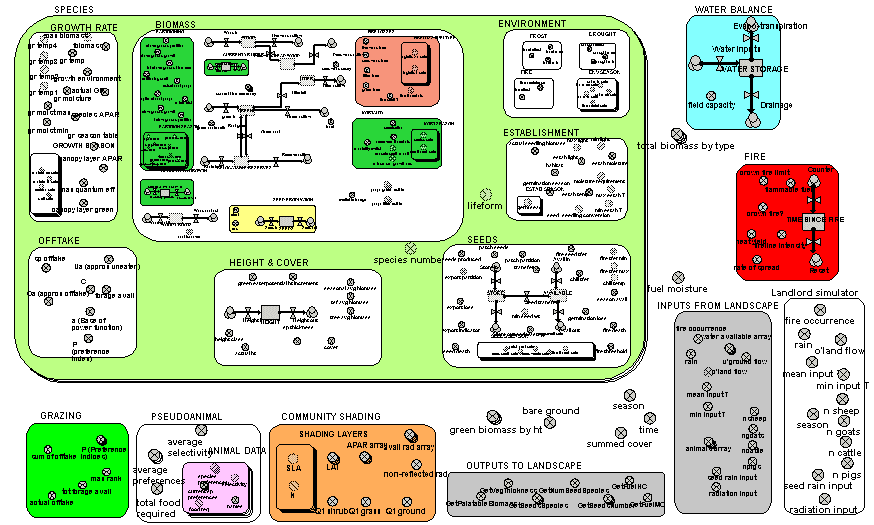
The preceding example shows one way in which the modelling of a vegetation component can be made more detailed – in that case, by disaggregating one component (the crown) into separate bits. This is appropriate for big, discrete vegetation components, such as trees. Another way of attempting to make a model more realistic is to model the processes themselves in more detail. The complete model diagram shows a multiple-species model of biomass dynamics developed using Simile in ModMED.
The multiple-boundary for the submodel SPECIES shows visually that several species are being modelled. Some components of the system (such as soil water balance and fire) are outside this submodel, indicating that they apply at the community level, rather than separately for each species.
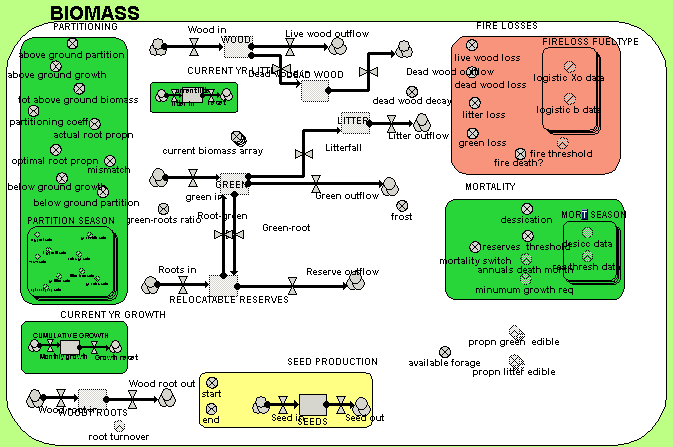
A zoomed-in view of the BIOMASS submodel enables the names of the individual variables to be seen.
In both diagrams, the influence arrows have been suppressed, for clarity. Simile allows users to choose which model diagram elements (including variables, flows and compartments) are displayed.
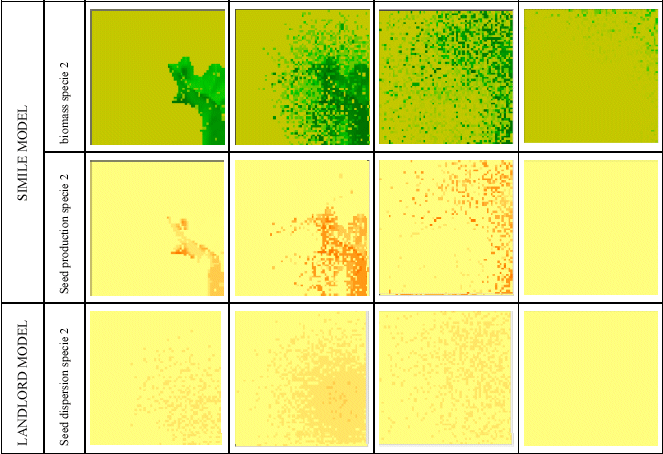
Seed dispersal using a GIS-based spatial modelling environment
Simile is capable of simulating both within-patch dynamics (e.g. vegetation dynamics) and between-patch dynamics (e.g. seed dispersal). However, Simile was introduced into the ModMED project in the third phase, after a GIS-based landscape-level modelling framework (called Landlord) had been developed. It was therefore decide to use Simile just for modelling at the patch (community) level, and to let LANDLORD manage seed dispersal, water movement and the spread of fire across the landscape.
This required that a Simile model could be incorporated into Landlord, and replicated across all the landscape grid cells. This was achieved by developing an interface which allowed the modeller to indicate the correspondence between variables in the Simile model and those in the landscape-level framework, and by generating a DLL for the Simile model which can be called by Landlord.
The following model diagram shows a simple biomass-based model of vegetation dynanics for several species. Note that it includes a seed production component.
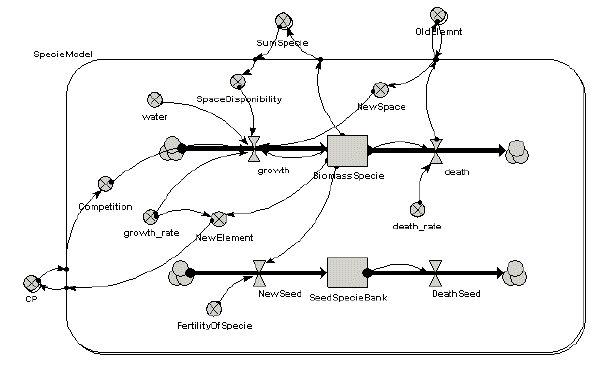
This model was then linked up to Landlord. These maps show the biomass and seed production simulated by the Simile model across all the grid cells of the landscape, at four points in time. The third row shows the seed dispersal calculated by Landlord, based on the seed production in each cell. This allows the vegetation to spread across the landscape over time.